Exploring the understudied human kinome for research and therapeutic opportunities
New work with Peter Sorger and Natarajan Kannan and led by Nienke Moret is now out on bioRxiv. (https://www.biorxiv.org/content/10.1101/2020.02.04.934216v1)
Abstract
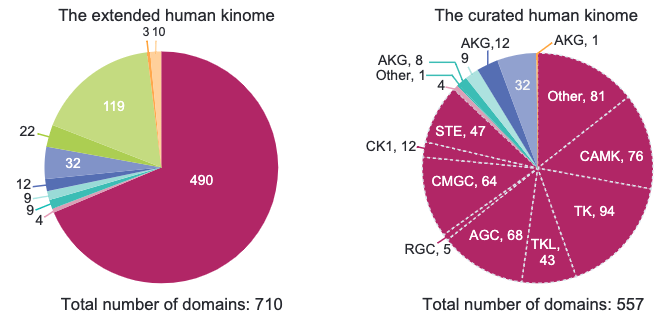
The functions of protein kinases have been heavily studied and inhibitors for many human kinases have been developed into FDA-approved therapeutics. A substantial fraction of the human kinome is nonetheless understudied. In this paper, members of the NIH Understudied Kinome Consortium mine public data on “dark” kinases to estimate the likelihood that they are functional. We start with a re-analysis of the human kinome and describe the criteria for creation of an inclusive set of 710 kinase domains and a curated set of 557 protein kinase like (PKL) domains. Nearly all PKLs are expressed in one or more CCLE cell lines and a substantial number are also essential in the Cancer Dependency Map. Dark kinases are frequently differentially expressed or mutated in The Cancer Genome Atlas and other disease databases and investigational and approved kinase inhibitors appear to inhibit them as off-target activities. Thus, it seems likely that the dark human kinome contains multiple biologically important genes, a subset of which may be viable drug targets.
Moret N, Liu C, Gyori BM, Bachman JA, Steppi A, Taujale R, Huang L-C, Hug C, Berginski M, Gomez S, Kannan N, Sorger PK. 2020. Exploring the understudied human kinome for research and therapeutic opportunities. bioRxiv:2020.04.02.022277. DOI: 10.1101/2020.04.02.022277.